fillNodata() is a wrapper for GDALFillNodata() in the GDAL Algorithms
API. This algorithm will interpolate values for all designated nodata
pixels (pixels having an intrinsic nodata value, or marked by zero-valued
pixels in the optional raster specified in mask_file). For each nodata
pixel, a four direction conic search is done to find values to interpolate
from (using inverse distance weighting).
Once all values are interpolated, zero or more smoothing iterations
(3x3 average filters on interpolated pixels) are applied to smooth out
artifacts.
Usage
fillNodata(
filename,
band,
mask_file = "",
max_dist = 100,
smooth_iterations = 0L,
quiet = FALSE
)Arguments
- filename
Filename of input raster in which to fill nodata pixels.
- band
Integer band number to modify in place.
- mask_file
Optional filename of raster to use as a validity mask (band 1 is used, zero marks nodata pixels, non-zero marks valid pixels).
- max_dist
Maximum distance (in pixels) that the algorithm will search out for values to interpolate (100 pixels by default).
- smooth_iterations
The number of 3x3 average filter smoothing iterations to run after the interpolation to dampen artifacts (0 by default).
- quiet
Logical scalar. If
TRUE, a progress bar will not be displayed. Defaults toFALSE.
Note
The input raster will be modified in place. It should not be open in a
GDALRaster object while processing with fillNodata().
Examples
## fill nodata edge pixels
f <- system.file("extdata/storml_elev_orig.tif", package="gdalraster")
## get count of nodata
tbl <- buildRAT(f)
#> scanning raster...
#> 0...10...20...30...40...50...60...70...80...90...100 - done.
head(tbl)
#> VALUE COUNT
#> 1 2438 9
#> 2 2439 6
#> 3 2440 5
#> 4 2441 5
#> 5 2442 5
#> 6 2443 2
tbl[is.na(tbl$VALUE),]
#> VALUE COUNT
#> 601 NA 876
ds <- new(GDALRaster, f)
plot_raster(ds, legend = TRUE)
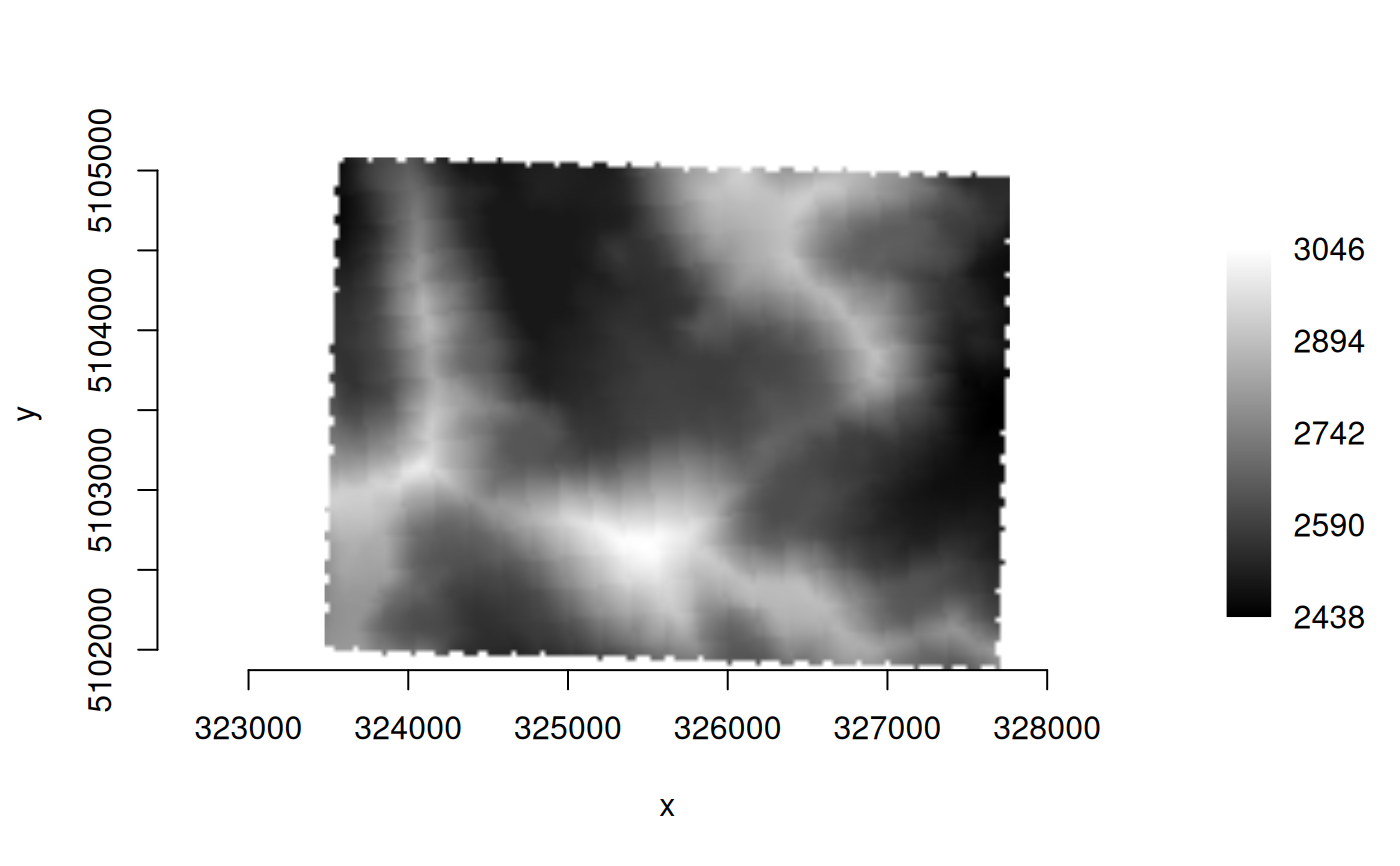 ds$close()
## make a copy that will be modified
mod_file <- file.path(tempdir(), "storml_elev_fill.tif")
file.copy(f, mod_file)
#> [1] TRUE
fillNodata(mod_file, band = 1)
#> 0...10...20...30...40...50...60...70...80...90...100 - done.
mod_tbl = buildRAT(mod_file)
#> scanning raster...
#> 0...10...20...30...40...50...60...70...80...90...100 - done.
head(mod_tbl)
#> VALUE COUNT
#> 1 2438 9
#> 2 2439 7
#> 3 2440 8
#> 4 2441 5
#> 5 2442 7
#> 6 2443 2
mod_tbl[is.na(mod_tbl$VALUE),]
#> [1] VALUE COUNT
#> <0 rows> (or 0-length row.names)
ds <- new(GDALRaster, mod_file)
plot_raster(ds, legend = TRUE)
ds$close()
## make a copy that will be modified
mod_file <- file.path(tempdir(), "storml_elev_fill.tif")
file.copy(f, mod_file)
#> [1] TRUE
fillNodata(mod_file, band = 1)
#> 0...10...20...30...40...50...60...70...80...90...100 - done.
mod_tbl = buildRAT(mod_file)
#> scanning raster...
#> 0...10...20...30...40...50...60...70...80...90...100 - done.
head(mod_tbl)
#> VALUE COUNT
#> 1 2438 9
#> 2 2439 7
#> 3 2440 8
#> 4 2441 5
#> 5 2442 7
#> 6 2443 2
mod_tbl[is.na(mod_tbl$VALUE),]
#> [1] VALUE COUNT
#> <0 rows> (or 0-length row.names)
ds <- new(GDALRaster, mod_file)
plot_raster(ds, legend = TRUE)
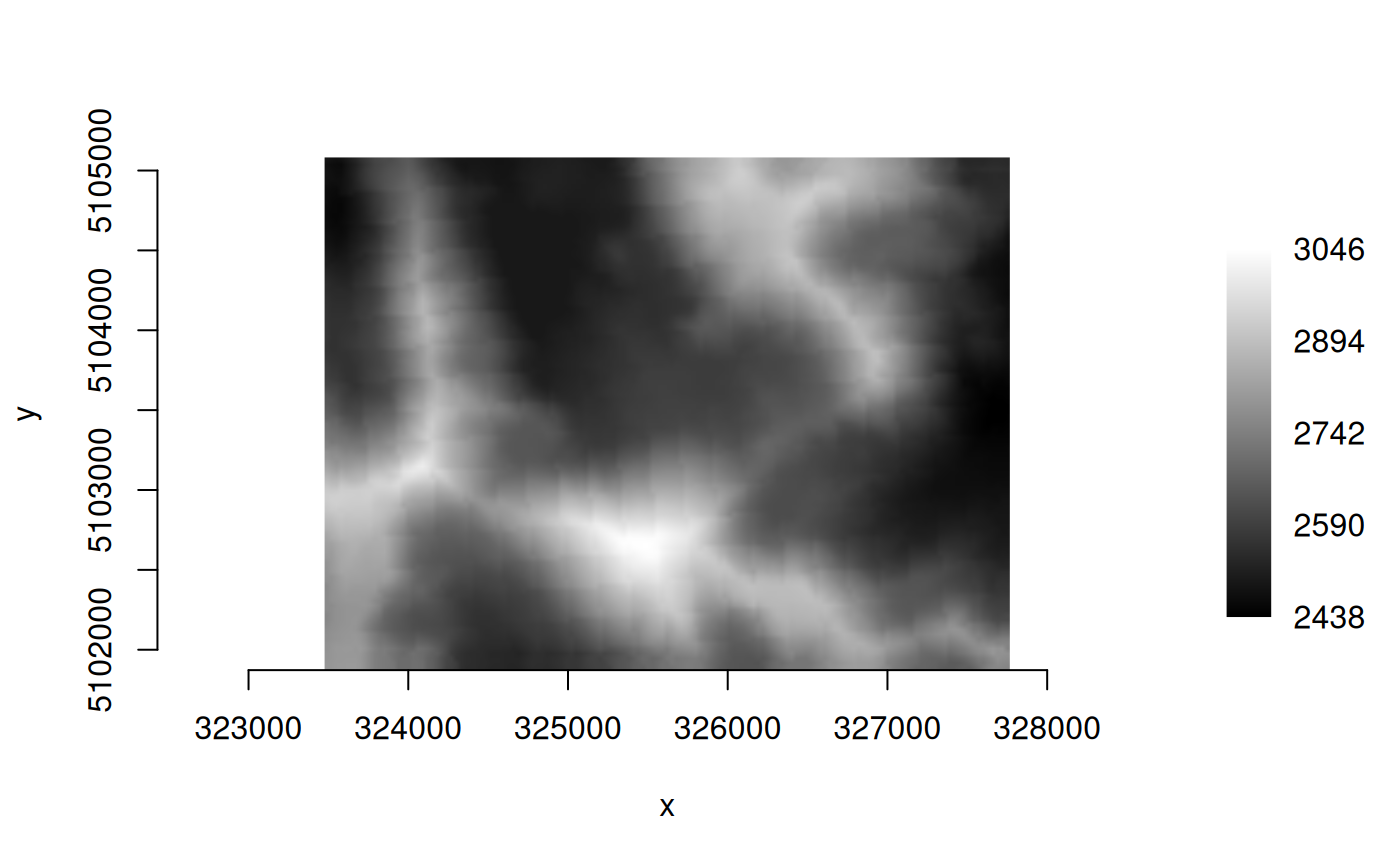 ds$close()
ds$close()